CSS Forums
Thursday, April 25, 2024
10:27 PM (GMT +5)
10:27 PM (GMT +5)
|
#111
|
|||
|
|||
|
|
|
#112
|
|||
|
|||
|
Endoplasmic Reticulum The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). OR The Endoplasmic Reticulum (ER) organelle is a network of disk-like tubules, sacks and vesicles found in eukaryotic cells. Its main function is to operate as a transport system. Endoplasmic reticulum is chiefly made up of a phospholipid membrane containing an internal fluid-filled space called the cisternal space or the lumen. The ER is folded and stacked layer upon layer within the cell and is connected to the cell's nuclear membrane. Under a microscope, the endoplasmic reticulum is seen as a highly folded structure surrounding the cell nucleus. There are two types of ER, Rough and Smooth. 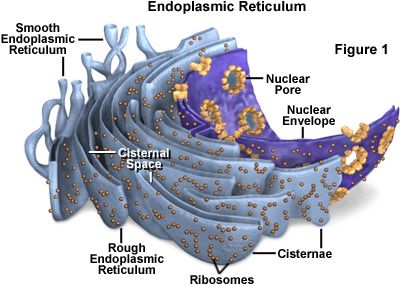 Structure. The general structure of the endoplasmic reticulum is an extensive membrane network of cisternae (sac-like structures) held together by the cytoskeleton. The phospholipid membrane encloses a space, the cisternal space (or lumen), from the cytosol. The functions of the endoplasmic reticulum vary greatly depending on the exact type of endoplasmic reticulum and the type of cell in which it resides. The three varieties are called rough endoplasmic reticulum, smooth endoplasmic reticulum and sarcoplasmic reticulum. The quantity of RER and SER in a cell can quickly interchange from one type to the other, depending on changing metabolic needs: one type will undergo numerous changes including new proteins embedded in the membranes in order to transform. Also, massive changes in the protein content can occur without any noticeable structural changes, depending on the enzymatic needs of the cell Rough Endoplasmic Reticulum Rough ER contains ribosomes on its surface, small circular structures that control protein synthesis, making it look bumpy under a microscope. Rough endoplasmic reticulum branches out and expands as protein synthesis increases, providing more surface area for ribosomes to spread out and create more proteins. During protein synthesis, the ribosomes on the rough ER create new proteins and the ER then folds them properly and sorts them according to function and destination. The rough ER carries out its protein folding activity deep within the cisternal space. There, newly made proteins are twisted and folded into the configurations necessary for them to carry out their function in the cell. For sorting, the rough ER works in conjunction with the Golgi apparatus, another cell organelle, to target the newly synthesized proteins to their proper locations. Most proteins produced by ribosomes of the rough endoplasmic reticulum are destined for secretion out of the cell. The RER is key in multiple functions:
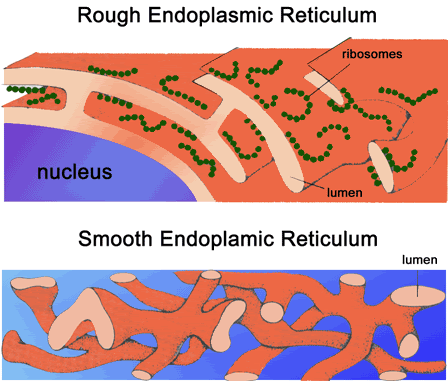 Smooth Endoplasmic Reticulum Smooth ER has a few different functions in the cell, and its functions can vary with cell type. In general, the function of smooth endoplasmic reticulum is to provide surface area for the action of enzymes and to provide storage space for these important enzymes. In muscle cells, for example, smooth ER stores calcium, releasing it during muscle contraction, triggering the movement of the muscle. In the liver, smooth ER is where detoxification takes place. Smooth ER also synthesizes lipids (fats) and other components of the cell membrane. Another function of smooth endoplasmic reticulum is to control the movement of newly synthesized proteins to their proper location in the cell or to the membrane to be sent outside the cell. A process called budding, where small vesicles of smooth ER are pinched off to carry the proteins to their new location, does this. The many functions of endoplasmic reticulum make it an important organelle for maintaining normal cell functioning. Defective or stressed endoplasmic reticulum has been implicated in diseases ranging from rare genetic conditions to well known conditions such as Alzheimer's disease, diabetes and heart disease. The Smooth ER also contains the enzyme glucose-6-phosphatase which converts glucose-6-phosphate to glucose, a step in gluconeogenesis.  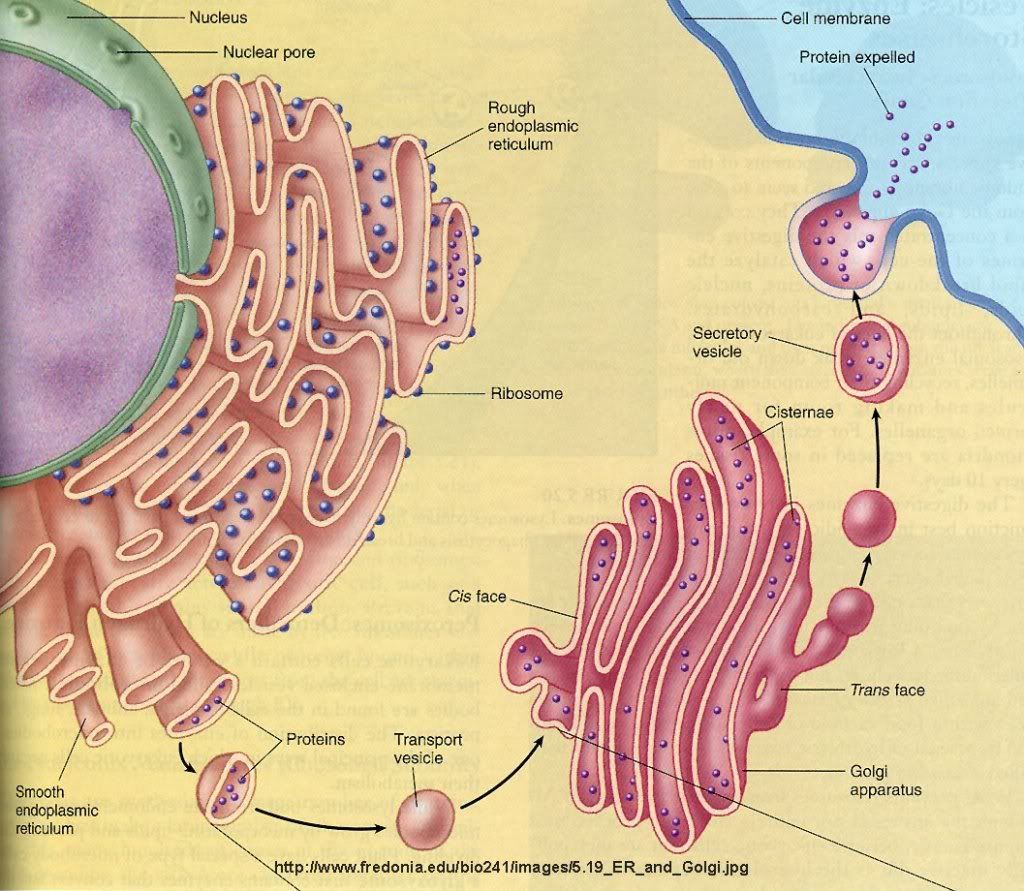 Functions The endoplasmic reticulum serves many general functions, including the facilitation of protein folding and the transport of synthesized proteins in sacs called cisternae. Correct folding of newly-made proteins is made possible by several endoplasmic reticulum chaperone proteins, including protein disulfide isomerase (PDI), ERp29, the Hsp70 family member Grp78, calnexin, calreticulin, and the peptidylpropyl isomerase family. Only properly-folded proteins are transported from the rough ER to the Golgi complex. Transport of proteins Secretory proteins, mostly glycoproteins, are moved across the endoplasmic reticulum membrane. Proteins that are transported by the endoplasmic reticulum and from there throughout the cell are marked with an address tag called a signal sequence. The N-terminus (one end) of a polypeptide chain (i.e., a protein) contains a few amino acids that work as an address tag, which are removed when the polypeptide reaches its destination. Proteins that are destined for places outside the endoplasmic reticulum are packed into transport vesicles and moved along the cytoskeleton toward their destination. The endoplasmic reticulum is also part of a protein sorting pathway. It is, in essence, the transportation system of the eukaryotic cell. The majority of endoplasmic reticulum resident proteins are retained in the endoplasmic reticulum through a retention motif. This motif is composed of four amino acids at the end of the protein sequence. The most common retention sequence is KDEL (lys-asp-glu-leu). However, variation on KDEL does occur and other sequences can also give rise to endoplasmic reticulum retention. It is not known if such variation can lead to sub-endoplasmic reticulum localizations. There are three KDEL receptors in mammalian cells, and they have a very high degree of sequence identity. The functional differences between these receptors remain to be established. Other functions
|
| The Following User Says Thank You to AFRMS For This Useful Post: | ||
dr.atifrana (Monday, June 08, 2009) | ||
|
#113
|
|||
|
|||
|
Ribosomes Ribosomes (from ribonucleic acid and "Greek: soma (meaning body)") are complexes of RNA and protein that are found in all cells. Ribosomes from bacteria, archaea and eukaryotes, the three domains of life on Earth, have significantly different structure and RNA. The ribosomes in the mitochondria of eukaryotic cells resemble those in bacteria, reflecting the evolutionary origin of this organelle. The ribosome functions in the expression of the genetic code from nucleic acid into protein, in a process called translation. Ribosomes do this by catalyzing the assembly of individual amino acids into polypeptide chains; this involves binding a messenger RNA and then using this as a template to join together the correct sequence of amino acids. This reaction uses adapters called transfer RNA molecules, which read the sequence of the messenger RNA and are attached to the amino acids. Ribosomes are about 20nm (200 Ångström) in diameter and are composed of 65% ribosomal RNA and 35% ribosomal proteins (known as a Ribonucleoprotein or RNP). They translate messenger RNA (mRNA) to build polypeptide chains (e.g., proteins) using amino acids delivered by transfer RNA (tRNA). Their active sites are made of RNA, so ribosomes are now classified as "ribozymes". 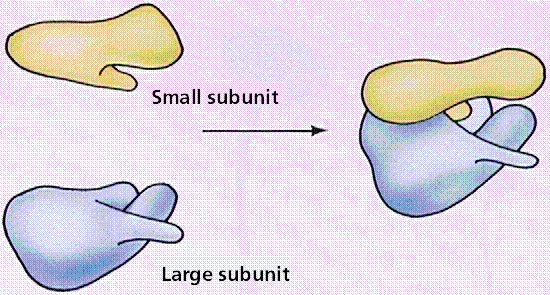 Ribosomes build proteins from the genetic instructions held within messenger RNA. Free ribosomes are suspended in the cytosol (the semi-fluid portion of the cytoplasm); others are bound to the rough endoplasmic reticulum, giving it the appearance of roughness and thus its name, or to the nuclear envelope. As ribozymes are partly constituted from RNA, it is thought that they might be remnants of the RNA world. Catalysis of the peptide bond involves the C2 hydroxyl of RNA's P-site adenosine in a protein shuttle mechanism. The full function (i.e. translocation) of the ribosome is reliant on changes in protein conformations. Ribosomes are sometimes referred to as organelles, but the use of the term organelle is often used only in reference to sub-cellular components that include a phospholipid membrane, which ribosomes, being entirely particulate, do not. For this reason, ribosomes may sometimes be described as "non-membranous organelles". 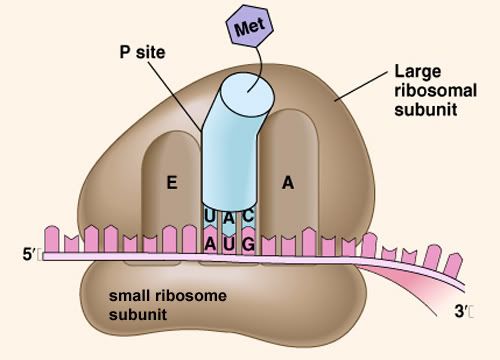 Ribosomes were first observed in the mid-1950s by Romanian cell biologist George Palade using an electron microscope as dense particles or granules for which he would win the Nobel Prize. The term "ribosome" was proposed by scientist Richard B. Roberts in 1958: Ribosomes consist of two subunits that fit together and work as one to translate the mRNA into a polypeptide chain during protein synthesis . Bacterial subunits consist of one or two and eukaryotic of one or three very large RNA molecules (known as ribosomal RNA or rRNA) and multiple smaller protein molecules. Crystallographic work has shown that there are no ribosomal proteins close to the reaction site for polypeptide synthesis. This suggests that the protein components of ribosomes act as a scaffold that may enhance the ability of rRNA to synthesize protein rather than directly participating in catalysis . Ribosome locations Ribosomes are classified as being either "free" or "membrane-bound." Free and membrane-bound ribosomes differ only in their spatial distribution; they are identical in structure and function. Whether the ribosome exists in a free or membrane-bound state depends on the presence of an ER-targeting signal sequence on the protein being synthesized. Free ribosomes Free ribosomes are free to move about anywhere in the cytosol. Proteins that are formed from free ribosomes are used within the cell. Proteins containing disulfide bonds using cysteine amino acids cannot be produced outside of the lumen of the endoplasmic reticulum. Membrane-bound ribosomes When certain proteins are synthesized by a ribosome they can become "membrane-bound". The newly produced polypeptide chains are inserted directly into the endoplasmic reticulum by the ribosome and are then transported to their destinations. Bound ribosomes usually produce proteins that are used within the cell membrane or are expelled from the cell via exocytosis Function Ribosomes are the workhorses of protein biosynthesis, the process of translating mRNA into protein. The mRNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon (3 nucleotides) of the mRNA, pairing it with the appropriate amino acid provided by a tRNA. Molecules of transfer RNA (tRNA) contain a complementary anticodon on one end and the appropriate amino acid on the other. The small ribosomal subunit, typically bound to a tRNA containing the amino acid methionine, binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. The ribosome then contains three RNA binding sites, designated A, P, and E. The A site binds an aminoacyl-tRNA (a tRNA bound to an amino acid); the P site binds a peptidyl-tRNA (a tRNA bound to the peptide being synthesized); and the E site binds a free tRNA before it exits the ribosome. Protein synthesis begins at a start codon AUG near the 5' end of the mRNA. mRNA binds to the P site of the ribosome first. The ribosome is able to identify the start codon by use of the Shine-Dalgarno sequence of the mRNA in prokaryotes and Kozak box in eukaryotes.  |
| The Following User Says Thank You to AFRMS For This Useful Post: | ||
dr.atifrana (Monday, June 08, 2009) | ||
|
#114
|
|||
|
|||
|
Animal Cell Nucleus The nucleus is a highly specialized organelle that serves as the information and administrative center of the cell. This organelle has two major functions. It stores the cell's hereditary material, or DNA, and it coordinates the cell's activities, which include intermediary metabolism, growth, protein synthesis, and reproduction (cell division). Only the cells of advanced organisms, known as eukaryotes, have a nucleus. Generally there is only one nucleus per cell, but there are exceptions such as slime molds and the Siphonales group of algae. Simpler one-celled organisms (prokaryotes), like the bacteria and cyanobacteria, don't have a nucleus. In these organisms, all the cell's information and administrative functions are dispersed throughout the cytoplasm. 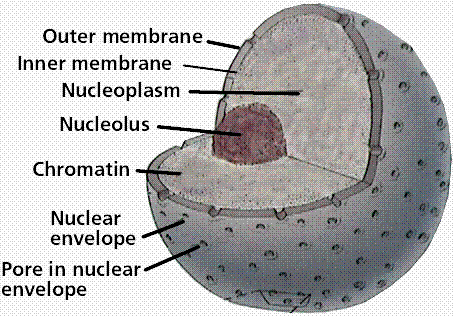 The spherical nucleus occupies about 10 percent of a cell's volume, making it the cell's most prominent feature. Most of the nuclear material consists of chromatin, the unstructured form of the cell's DNA that will organize to form chromosomes during mitosis or cell division. Also inside the nucleus is the nucleolus, an organelle that synthesizes protein-producing macromolecular assemblies called ribosomes. A double-layered membrane, the nuclear envelope, separates contents of the nucleus from the cellular cytoplasm. The envelope is riddled with holes called nuclear pores that allow specific types and sizes of molecules to pass back and forth between the nucleus and the cytoplasm. It is also attached to a network of tubules, called the endoplasmic reticulum, where protein synthesis occurs. These tubules extend throughout the cell and manufacture the biochemical products that a particular cell type is genetically coded to produce. · Chromatin/Chromosomes - Packed inside the nucleus of every human cell is nearly 6 feet of DNA, which is divided into 46 individual molecules, one for each chromosome and each about 1.5 inches long. Packing all this material into a microscopic cell nucleus is an extraordinary feat of packaging. For DNA to function, it can't be crammed into the nucleus like a ball of string. Instead, it is combined with proteins and organized into a precise, compact structure, a dense string-like fiber called chromatin. Each DNA strand wraps around groups of small protein molecules called histones, forming a series of bead-like structures, called nucleosomes, connected by the DNA strand. Under the microscope, uncondensed chromatin has a "beads on a string" appearance. The string of nucleosomes, already compacted by a factor of six, is then coiled into an even denser structure, compacting the DNA by a factor of 40. This compression and structuring of DNA serves several functions. The overall negative charge of the DNA is neutralized by the positive charge of the histone molecules, the DNA takes up much less space, and inactive DNA can be folded into inaccessible locations until it is needed. There are two types of chromatin. Euchromatin is the genetically active portion and is involved in transcribing RNA to produce proteins used in cell function and growth. Heterochromatin contains inactive DNA and is the portion of chromatin that is most condensed, since it not being used. Throughout the life of a cell, chromatin fibers take on different forms inside the nucleus. During interphase, when the cell is carrying out its normal functions, the chromatin is dispersed throughout the nucleus in what appears to be a tangle of fibers. This exposes the euchromatin and makes it available for the transcription process. When the cell enters metaphase and prepares to divide, the chromatin changes dramatically. First, all the chromatin strands make copies of themselves through the process of DNA replication. Then they are compressed to an even greater degree than at interphase, a 10,000-fold compaction, into specialized structures for reproduction, termed chromosomes. As the cell divides to become two cells, the chromosomes separate, giving each cell a complete copy of the genetic information contained in the chromatin. · Nucleolus - The nucleolus is a membrane-less organelle within the nucleus that manufactures ribosomes, the cell's protein-producing structures. Through the microscope, the nucleolus looks like a large dark spot within the nucleus. A nucleus may contain up to four nucleoli, but within each species the number of nucleoli is fixed. After a cell divides, a nucleolus is formed when chromosomes are brought together into nucleolar organizing regions. During cell division, the nucleolus disappears. Some studies suggest that the nucleolus may be involved with cellular aging and, therefore, may affect the aging of an organism. · Nuclear Envelope - The nuclear envelope is a double-layered membrane that encloses the contents of the nucleus during most of the cell’s lifecycle. The space between the layers is called the perinuclear space and appears to connect with the rough endoplasmic reticulum. The envelope is perforated with tiny holes called nuclear pores. These pores regulate the passage of molecules between the nucleus and cytoplasm, permitting some to pass through the membrane, but not others. The inner surface has a protein lining called the nuclear lamina, which binds to chromatin and other nuclear components. During mitosis, or cell division, the nuclear envelope disintegrates, but reforms as the two cells complete their formation and the chromatin begins to unravel and disperse. 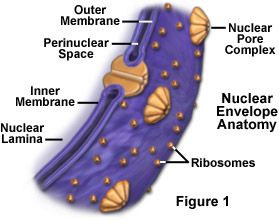 · Nuclear Pores - The nuclear envelope is perforated with holes called nuclear pores. These pores regulate the passage of molecules between the nucleus and cytoplasm, permitting some to pass through the membrane, but not others. Building blocks for building DNA and RNA are allowed into the nucleus as well as molecules that provide the energy for constructing genetic material. The pores are fully permeable to small molecules up to the size of the smallest proteins, but form a barrier keeping most large molecules out of the nucleus. Some larger proteins, such as histones, are given admittance into the nucleus. Each pore is surrounded by an elaborate protein structure called the nuclear pore complex, which probably selects large molecules for entrance into the nucleus. |
| The Following User Says Thank You to AFRMS For This Useful Post: | ||
dr.atifrana (Monday, June 08, 2009) | ||
|
#115
|
|||
|
|||
|
Golgi Apparatus The Golgi apparatus (GA), also called Golgi body or Golgi complex and found universally in both plant and animal cells, is typically comprised of a series of five to eight cup-shaped, membrane-covered sacs called cisternae that look something like a stack of deflated balloons. In some unicellular flagellates, however, as many as 60 cisternae may combine to make up the Golgi apparatus. Similarly, the number of Golgi bodies in a cell varies according to its function. Animal cells generally contain between ten and twenty Golgi stacks per cell, which are linked into a single complex by tubular connections between cisternae. This complex is usually located close to the cell nucleus. Discovery Due to its relatively large size, the Golgi apparatus was one of the first organelles ever observed. In 1897, an Italian physician named Camillo Golgi, who was investigating the nervous system by using a new staining technique he developed (and which is still sometimes used today; known as Golgi staining or Golgi impregnation), observed in a sample under his light microscope a cellular structure that he termed the internal reticular apparatus. Soon after he publicly announced his discovery in 1898, the structure was named after him, becoming universally known as the Golgi apparatus. Yet, many scientists did not believe that what Golgi observed was a real organelle present in the cell and instead argued that the apparent body was a visual distortion caused by staining. The invention of the electron microscope in the twentieth century finally confirmed that the Golgi apparatus is a cellular organelle. 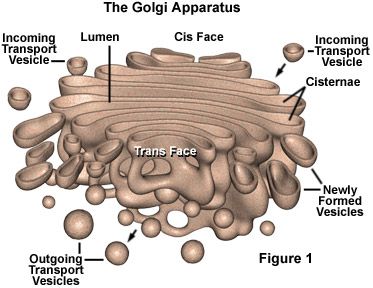 Structure In general, the Golgi apparatus is made up of approximately four to eight cisternae, although in some single-celled organisms it may consist of as many as 60 cisternae. Matrix proteins hold the cisternae together, and the whole of the Golgi apparatus is supported by cytoplasmic microtubules. The apparatus has three primary compartments, known generally as “cis” (cisternae nearest the endoplasmic reticulum), “medial” (central layers of cisternae), and “trans” (cisternae farthest from the endoplasmic reticulum). Two networks, the cis Golgi network and the trans Golgi network, which are made up of the outermost cisternae at the cis and trans faces, are responsible for the essential task of sorting proteins and lipids that are received (at the cis face) or released (at the trans face) by the organelle. Transport of substances The proteins and lipids received at the cis face arrive in clusters of fused vesicles. These fused vesicles migrate along microtubules through a special trafficking compartment, called the vesicular-tubular cluster that lies between the endoplasmic reticulum and the Golgi apparatus. When a vesicle cluster fuses with the cis membrane, the contents are delivered into the lumen of the cis face cisterna. As proteins and lipids progress from the cis face to the trans face, they are modified into functional molecules and are marked for delivery to specific intracellular or extra cellular locations. Some modifications involve cleavage of oligosaccharide side chains followed by attachment of different sugar moieties in place of the side chain. Other modifications may involve the addition of fatty acids or phosphate groups (phosphorylation) or the removal of monosaccharides. The different enzyme driven modification reactions are specific to the compartments of the Golgi apparatus. For example, the removal of mannose moieties occurs primarily in the cis and medial cisternae, whereas the addition of galactose or sulfate occurs primarily in the trans cisternae. In the final stage of transport through the Golgi apparatus, modified proteins and lipids are sorted in the trans Golgi network and are packaged into vesicles at the trans face. These vesicles then deliver the molecules to their target destinations, such as lysosomes or the cell membrane. Some molecules, including certain soluble proteins and secretory proteins, are carried in vesicles to the cell membrane for exocytosis (release into the extracellular environment). The exocytosis of secretory proteins may be regulated, whereby a ligand must bind to a receptor to trigger vesicle fusion and protein secretion. 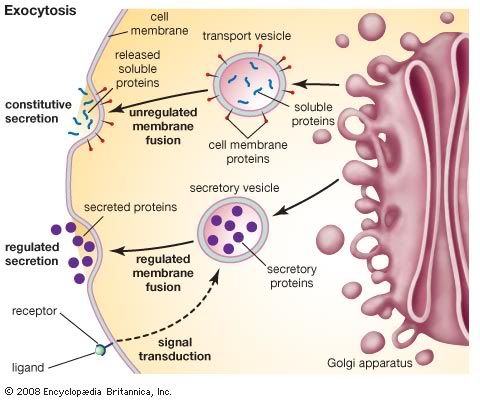 The way in which proteins and lipids move from the cis face to the trans face is of some debate, and today there exist two models, with quite different perceptions of the Golgi apparatus, competing to explain this movement. The vesicular transport model stems from initial studies that identified vesicles in association with the Golgi apparatus. This model is based on the idea that vesicles bud off and fuse to cisternae membranes, thus moving molecules from one cisterna to the next; budding vesicles can also be used to transport molecules back to the endoplasmic reticulum. A vital element of this model is that the cisternae themselves are stationary. In contrast, the cisternal maturation model depicts the Golgi apparatus as a far more dynamic organelle than does the vesicular transport model. The cisternal maturation model indicates that cis cisternae move forward and mature into trans cisternae, with new cis cisternae forming from the fusion of vesicles at the cis face. In this model, vesicles are formed but are used only to transport molecules back to the endoplasmic reticulum. |
| The Following User Says Thank You to AFRMS For This Useful Post: | ||
dr.atifrana (Monday, June 08, 2009) | ||
|
#116
|
|||
|
|||
|
Mitochondria In cell biology, a mitochondrion (plural mitochondria) is a membrane-enclosed organelle found in most eukaryotic cells. These organelles range from 0.5–10 micrometers (μm) in diameter. Mitochondria are sometimes described as "cellular power plants" because they generate most of the cell's supply of adenosine triphosphate(ATP), used as a source of chemical energy In addition to supplying cellular energy, mitochondria are involved in a range of other processes, such as signaling, cellular differentiation, cell death, as well as the control of the cell cycle and cell growth Mitochondria have been implicated in several human diseases, including mitochondrial disorders and cardiac dysfunction,and may play a role in the aging process. Several characteristics make mitochondria unique. The number of mitochondria in a cell varies widely by organism and tissue type. Many cells have only a single mitochondrion, whereas others can contain several thousand mitochondria.The organelle is composed of compartments that carry out specialized functions. These compartments or regions include the outer membrane, the intermembrane space, the inner membrane, and the cristae and matrix. Mitochondrial proteins vary depending on the tissues and species. In human, 615 distinct types of proteins were identified from cardiac mitochondria, whereas in murinae (rats), 940 proteins encoded by distinct genes were reported.The mitochondrial proteome is thought to be dynamically regulated. Although most of a cell's DNA is contained in the cell nucleus, the mitochondrion has its own independent genome. Further, its DNA shows substantial similarity to bacterial genomes. 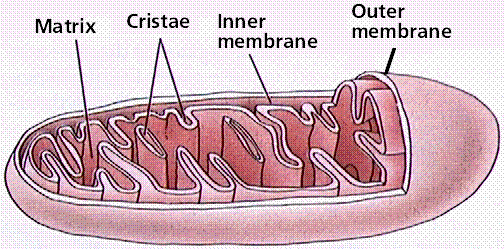 Structure A mitochondrion contains outer and inner membranes composed of phospholipid bilayers and proteins. The two membranes, however, have different properties. Because of this double-membraned organization, there are five distinct compartments within the mitochondrion. There is the outer mitochondrial membrane, the intermembrane space (the space between the outer and inner membranes), the inner mitochondrial membrane, the cristae space (formed by infoldings of the inner membrane), and the matrix(space within the inner membrane). Outer mitochondrial membrane The outer mitochondrial membrane, which encloses the entire organelle, has a protein-to-phospholipid ratio similar to that of the eukaryotic plasma membrane (about 1:1 by weight). It contains large numbers of integral proteins called porins.. These porins form channels that allow molecules 5000 Daltons or less in molecular weight to freely diffuse from one side of the membrane to the other. Larger proteins can also enter the mitochondrion if a signaling sequence at their N-terminusbinds to a large multisubunit protein called translocase of the outer membrane, which then actively moves them across the membrane.Disruption of the outer membrane permits proteins in the intermembrane space to leak into the cytosol, leading to certain cell death. Intermembrane space The intermembrane space is basically the space between the outer membrane and the inner membrane. Because the outer membrane is freely permeable to small molecules, the concentrations of small molecules such as ions and sugars in the intermembrane space is the same as the cytosol.However, as large proteins must have a specific signaling sequence to be transported across the outer membrane, the protein composition of this space is different than the protein composition of the cytosol. One protein that is localized to the intermembrane space in this way is cytochrome c. Inner mitochondrial membrane The inner mitochondrial membrane contains proteins with four types of functions:
Cristae The inner mitochondrial membrane is compartmentalized into numerous cristae, which expand the surface area of the inner mitochondrial membrane, enhancing its ability to produce ATP. These are not simple random folds but rather invaginations of the inner membrane, which can affect overall chemiosmotic function.In typical livermitochondria, for example, the surface area, including cristae, is about five times that of the outer membrane. Mitochondria of cells that have greater demand for ATP, such as muscle cells, contain more cristae than typical liver mitochondria.These folds are studded with small round bodies known as F1 particles or oxysomes. Matrix The matrix is the space enclosed by the inner membrane. It contains about 2/3 of the total protein in a mitochondrion.The matrix is important in the production of ATP with the aid of the ATP synthase contained in the inner membrane. The matrix contains a highly-concentrated mixture of hundreds of enzymes, special mitochondrial ribosomes, tRNA, and several copies of the mitochondrial DNA genome. Of the enzymes, the major functions include oxidation of pyruvate and fatty acids, and the citric acid cycle. Mitochondria have their own genetic material, and the machinery to manufacture their own RNAsand proteins. A published human mitochondrial DNA sequence revealed 16,569 base pairs encoding 37 total genes: 22 tRNA, 2 rRNA, and 13 peptide genes.The 13 mitochondrial peptides in humans are integrated into the inner mitochondrial membrane, along with proteins encoded by genes that reside in the host cell's nucleus. 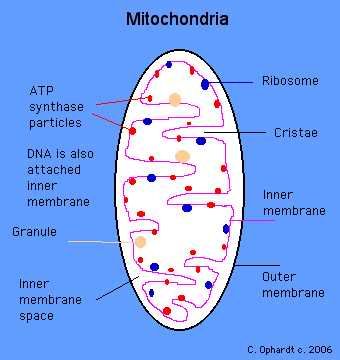 |
|
#117
|
|||
|
|||
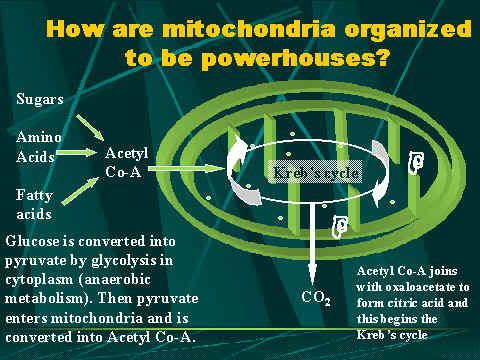 Steps from glycolysis to the electron transport chain Once inside the cell, glucose is broken down to make ATP in two pathways. The first pathway requires no oxygen and is called anaerobic metabolism. This pathway is called glycolysis and it occurs in the cytoplasm outside the mitochondria. During glycolysis, glucose is broken down into pyruvate. Other foods like fats can also be broken down for use as fuel. Each reaction is designed to produce some hydrogen ions (electrons) that can be used to make energy packets (ATP). However, only 4 ATP molecules can be made by one molecule of glucose run through this pathway. That is why mitochondria and oxygen are so important. We need to continue the breakdown process with the Krebs’s cycle inside the mitochondria in order to get enough ATP to run all the cell functions. 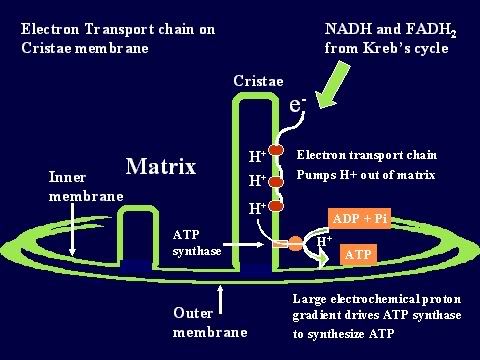 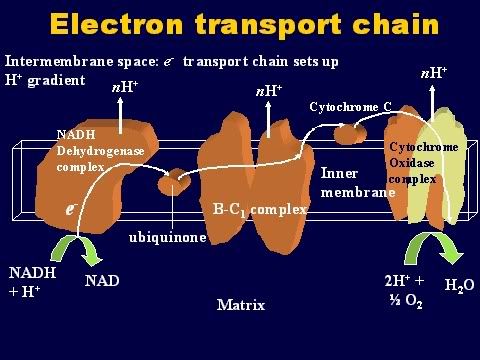 The whole idea behind respiration in the mitochondria is to use the Kreb’s cycle (also called the citric acid cycle) to get as many electrons out of the food we eat as possible. These electrons (in the form of hydrogen ions) are then used to drive pumps that produce ATP. The energy carried by ATP is then used for all kinds of cellular functions like movement, transport, entry and exit of products, division, etc. To run the Kreb's cycle, you need several important molecules in addition to all the enzymes. First; you need pyruvate, which is made by glycolysis from glucose. Next, you need some carrier molecules for the electrons. There are two types of these:
Oxidative phosphorylation First, some basic definitions. When you take hydrogen ions or electrons away from a molecule, you “oxidize” that molecule. When you give hydrogen ions or electrons to a molecule, you “reduce” that molecule. When you give phosphate molecules to a molecule, you “phosphorylate” that molecule. So, oxidative phosphorylation (very simply) means the process that couples the removal of hydrogen ions from one molecule and giving phosphate molecules to another molecule. As the Kreb’s cycle runs, hydrogen ions (or electrons) are donated to the two carrier molecules in 4 of the steps. Either NAD or FAD picks them up and these carrier molecules become NADH and FADH (because they now are carrying a hydrogen ion). These electrons are carried chemically to the respiratory or electron transport chain found in the mitochondrial cristae . The NADH and FADH essentially serve as a ferry in the lateral plane of the membrane diffusing from one complex to the next. At each site is a hydrogen (or proton) pumps, which transfers hydrogen from one side of the membrane to the other. This creates a gradient across the inner membrane with a higher concentration of Hydrogen ions in the intercristae space (this is the space between the inner and outer membranes). The electrons are carried from complex to complex by ubiquinone and cycochrome C. The third pump in the series catalyzes the transfer of the electrons to oxygen to make water. This chemiosmotic pumping creates an electrochemical proton gradient across the membrane, which is used to drive the “energy-producing machine”the ATP synthase. This molecule is found in small elementary particles that project from the cristae. As stated above, this process requires oxygen, which is why it is called "aerobic metabolism". The ATP synthase uses the energy of the hydrogen ion (also called proton) gradient to form ATP from ADP and Phosphate. It also produces water from the hydrogen and the oxygen. Thus, each compartment in the mitochondrion is specialized for one phase of these reactions. This is how oxidation is coupled to phosphorylation: NAD and FAD remove the electrons that are donated during some of the steps of the Kreb's or Citric acid cycle. Then, they carry the electrons to the electron transport pumps and donate them to the pumps. So, NAD and FAD are “oxidized” because they lose the hydrogen ions to the pumps. The pumps then transport the hydrogens ions to the space between the two membranes where they accumulate in a high enough concentration to fuel the ATP pumps. With sufficient fuel, they “phosphorylate” the ADP. That is how “oxidation” is coupled to “phosphorylation”. The hydrogens that get pumped back into the matrix by the ATP pump then combine with the oxygen to make water. And that is very important because, without oxygen, they will accumulate and the concentration gradient needed to run the ATP pumps will not allow the pumps to work. 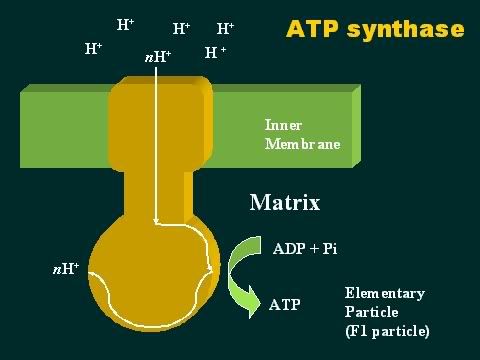 |
|
#118
|
|||
|
|||
|
Mitosis Mitosis is nuclear division plus cytokinesis, and produces two identical daughter cells during prophase, prometaphase, metaphase, anaphase, and telophase. Interphase is often included in discussions of mitosis, but interphase is technically not part of mitosis, but rather encompasses stages G1, S, and G2 of the cell cycle. Interphase The cell is engaged in metabolic activity and performing its prepare for mitosis (the next four phases that lead up to and include nuclear division). Chromosomes are not clearly discerned in the nucleus, although a dark spot called the nucleolus may be visible. The cell may contain a pair of centrioles (or microtubule organizing centers in plants) both of which are organizational sites for microtubules. Prophase Chromatin in the nucleus begins to condense and becomes visible in the light microscope as chromosomes. The nucleolus disappears. Centrioles begin moving to opposite ends of the cell and fibers extend from the centromeres. Some fibers cross the cell to form the mitotic spindle. Prometaphase The nuclear membrane dissolves, marking the beginning of prometaphase. Proteins attach to the centromeres creating the kinetochores. Microtubules attach at the kinetochores and the chromosomes begin moving. Metaphase Spindle fibers align the chromosomes along the middle of the cell nucleus. This line is referred to as the metaphase plate. This organization helps to ensure that in the next phase, when the chromosomes are separated, each new nucleus will receive one copy of each chromosome. Anaphase The paired chromosomes separate at the kinetochores and move to opposite sides of the cell. Motion results from a combination of kinetochore movement along the spindle microtubules and through the physical interaction of polar microtubules. Telophase Chromatids arrive at opposite poles of cell, and new membranes form around the daughter nuclei. The chromosomes disperse and are no longer visible under the light microscope. The spindle fibers disperse, and cytokinesis or the partitioning of the cell may also begin during this stage. Cytokinesis In animal cells, cytokinesis results when a fiber ring composed of a protein called actin around the center of the cell contracts pinching the cell into two daughter cells, each with one nucleus. In plant cells, the rigid wall requires that a cell plate be synthesized between the two daughter cells. 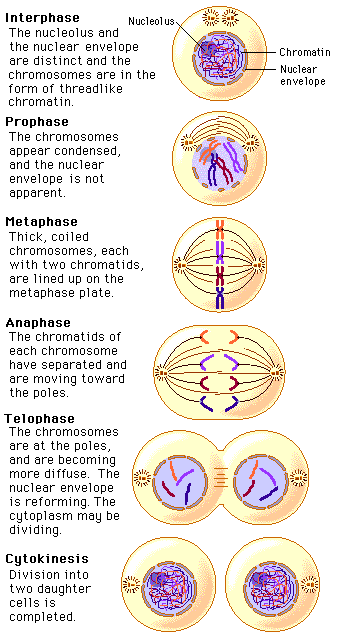 |
| The Following User Says Thank You to AFRMS For This Useful Post: | ||
paradox (Friday, August 07, 2009) | ||
|
#119
|
|||
|
|||
|
Meiosis Sexual reproduction occurs only in eukaryotes. During the formation of gametes,the number of chromosomes is reduced by half, and returned to the full amount when the two gametes fuse during fertilization. Ploidy Haploid and diploid are terms referring to the number of sets of chromosomes in a cell. Gregor Mendel determined his peas had two sets of alleles, one from each parent. Diploid organisms are those with two (di) sets. Human beings (except for their gametes), most animals and many plants are diploid. We abbreviate diploid as 2n. Ploidy is a term referring to the number of sets of chromosomes. Haploid organisms/cells have only one set of chromosomes, abbreviated, as n. Organisms with more than two sets of chromosomes are termed polyploid. Chromosomes that carry the same genes are termed homologous chromosomes. The alleles on homologous chromosomes may differ, as in the case of heterozygous individuals. Organisms (normally) receive one set of homologous chromosomes from each parent. Meiosis is a special type of nuclear division which segregates one copy of each homologous chromosome into each new "gamete". Mitosis maintains the cell's original ploidy level (for example, one diploid 2n cell producing two diploid 2n cells; one haploid n cell producing two haploid n cells; etc.). Meiosis, on the other hand, reduces the number of sets of chromosomes by half, so that when gametic recombination (fertilization)occurs the ploidy of the parents will be reestablished. Most cells in the human body are produced by mitosis. These are the somatic (or vegetative) line cells. Cells that become gametes are referred to as germ line cells. The vast majority of cell divisions in the human body are mitotic, with meiosis being restricted to the gonads. Phases of Meiosis Two successive nuclear divisions occur, Meiosis I (Reduction) and Meiosis II (Division). Meiosis produces 4 haploid cells. Mitosis produces 2 diploid cells. The old name for meiosis was reduction/ division. Meiosis I reduces the ploidy level from 2n to n (reduction) while Meiosis II divides the remaining set of chromosomes in a mitosis-like process (division). Most of the differences between the processes occur during Meiosis I. Prophase I Prophase I have a unique event -- the pairing (by an as yet undiscovered mechanism) of homologous chromosomes.Synapsis is the process of linking of the replicated homologous chromosomes. The resulting chromosome is termed a tetrad,being composed of two chromatids from each chromosome, forming a thick (4-strand) structure. Crossing-over may occur at this point. During crossing-over chromatids break and may be reattached to a different homologous chromosome. 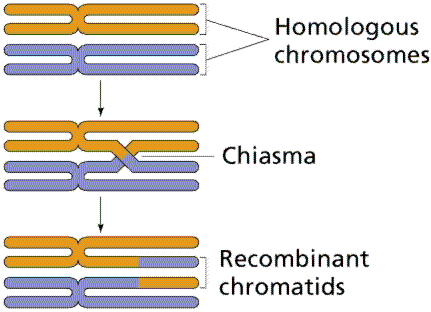 Events of Prophase I (save for synapsis and crossing over) are similar to those in Prophase of mitosis: chromatin condenses into chromosomes, the nucleolus dissolves, nuclear membrane is disassembled, and the spindle apparatus forms. 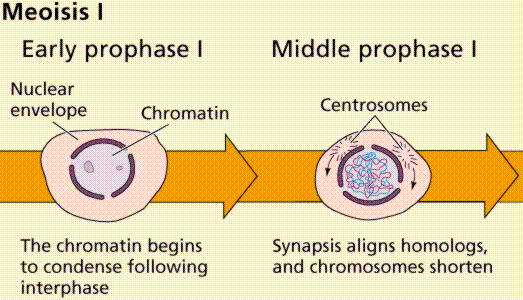 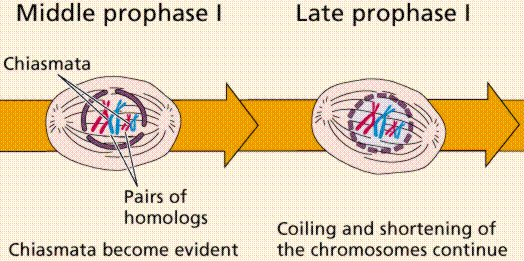 Metaphase I Metaphase I is when tetrads line-up along the equator of the spindle. Spindle fibers attach to the centromere region of each homologous chromosome pair. Other metaphase events as in mitosis. 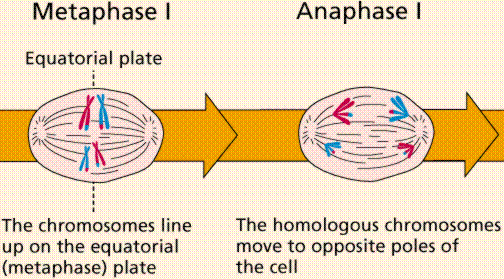 Anaphase I Anaphase I is when the tetrads separate, and are drawn to opposite poles by the spindle fibers. The centromeres in Anaphase I remain intact. Telophase I Telophase I is similar to Telophase of mitosis, except that only one set of (replicated) chromosomes is in each "cell". Depending on species, new nuclear envelopes may or may not form. Some animal cells may have division of the centrioles during this phase. 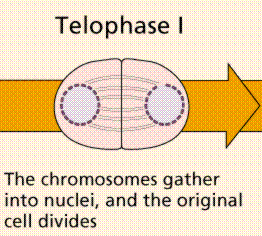 Meoisis II Prophase II During Prophase II, nuclear envelopes (if they formed during Telophase I) dissolve, and spindle fibers reform. All else is as in Prophase of mitosis. Indeed Meiosis II is very similar to mitosis. 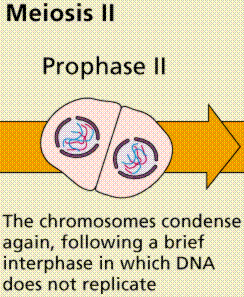 Metaphase II is similar to mitosis, with spindles moving chromosomes into equatorial area and attaching to the opposite sides of the centromeres in the kinetochore region. 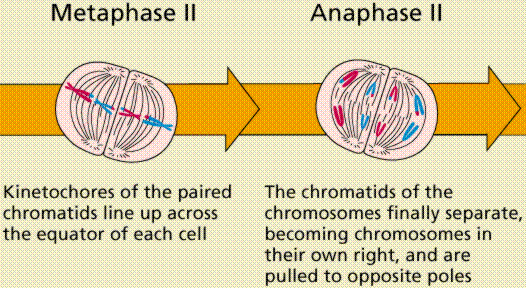 Anaphase II During Anaphase II, the centromeres split and the former chromatids (now chromosomes) are segregated into opposite sides of the cell. Telophase II Telophase II is identical to Telophase of mitosis. Cytokinesis separates the cells. 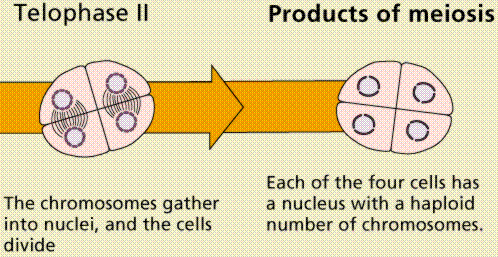 |
|
#120
|
|||
|
|||
|
DNA Replication. The process of making an identical copy of a section of duplex (double-stranded) DNA, using existing DNA as a template for the synthesis of new DNA strands. In humans and other eukaryotes, replication occurs in the cell nucleus. Before a cell can divide, it must duplicate all its DNA. In eukaryotes, this occurs during S phase of the cell cycle. The Biochemical Reactions DNA replication begins with the "unzipping" of the parent molecule as the hydrogen bonds between the base pairs are broken. Once exposed, the sequence of bases on each of the separated strands serves as a template to guide the insertion of a complementary set of bases on the strand being synthesized. The new strands are assembled from deoynucleotide triphosphate. Each incoming nucleotide is covalently linked to the "free" 3' carbon atom on the pentose (figure) as the second and third phosphates are removed together as a molecule of pyrophosphate(PPi). The nucleotides are assembled in the order that complements the order of bases on the strand serving as the template. Thus each C on the template guides the insertion of a G on the new strand, each G a C, and so on. When the process is complete, two DNA molecules have been formed identical to each other and to the parent molecule. The Enzymes A portion of the double helix is unwound by a helicase. A molecule of a DNA polymerase binds to one strand of the DNA and begins moving along it in the 3' to 5' direction, using it as a template for assembling a leading strand of nucleotides and reforming a double helix. In eukaryotes, this molecule is called DNA polymerase delta (δ). Because DNA synthesis can only occur 5' to 3', a molecule of a second type of DNA polymerase (epsilon, ε, in eukaryotes) binds to the other template strand as the double helix opens. This molecule must synthesize discontinuous segments of polynucleotides (called Okazaki fragments). Another enzyme, DNA ligase I then stitches these together into the lagging strand. DNA Replication is Semiconservative When the replication process is complete, two DNA molecules — identical to each other and identical to the original — have been produced. Each strand of the original molecule has remained intact as it served as the template for the synthesis of a complementary strand. This mode of replication is described as semi-conservative: one-half of each new molecule of DNA is old; one-half new. Watson and Crick had suggested that this was the way the DNA would turn out to be replicated. Proof of the model came from the experiments of Meselson and Stahl. 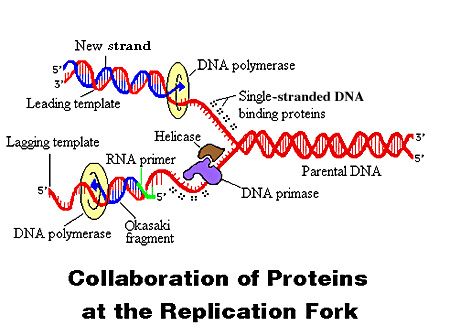 Steps of DNA Replication 1)The first major step for the DNA Replication to take place is the breaking of hydrogen bonds between bases of the two antiparallel strands. The unwounding of the two strands is the starting point. The splitting happens in places of the chains which are rich in A-T. That is because there are only two bonds between Adenine and Thymine (there are three hydrogen bonds between Cytosine and Guanine). Helicase is the enzyme that splits the two strands. The initiation point where the splitting starts is called "origin of replication".The structure that is created is known as "Replication Fork". 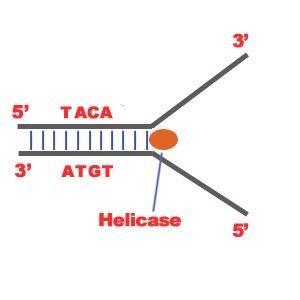 2) One of the most important steps of DNA Replication is the binding of RNA Primase in the the initiation point of the 3'-5' parent chain. RNA Primase can attract RNA nucleotides which bind to the DNA nucleotides of the 3'-5' strand due to the hydrogen bonds between the bases. RNA nucleotides are the primers (starters) for the binding of DNA nucleotides. 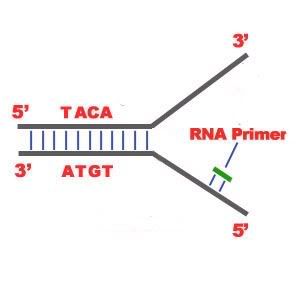 3) The elongation process is different for the 5'-3' and 3'-5' template. a)5'-3' Template: The 3'-5' proceeding daughter strand -that uses a 5'-3' template- is called leading strand because DNA Polymerase ä can "read" the template and continuously adds nucleotides (complementary to the nucleotides of the template, for example Adenine opposite to Thymine etc). b)3'-5'Template: The 3'-5' template cannot be "read" by DNA Polymerase ä. The replication of this template is complicated and the new strand is called lagging strand. In the lagging strand the RNA Primase adds more RNA Primers. DNA polymerase å reads the template and lengthens the bursts. The gap between two RNA primers is called "Okazaki Fragments". The RNA Primers are necessary for DNA Polymerase å to bind Nucleotides to the 3' end of them. The daughter strand is elongated with the binding of more DNA nucleotides. 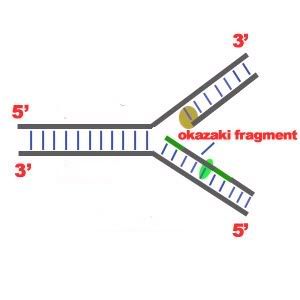 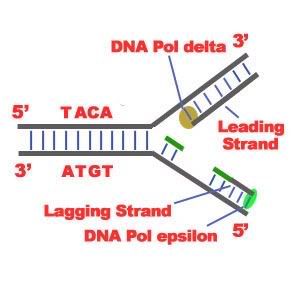 4) In the lagging strand the DNA Pol I -exonuclease- reads the fragments and removes the RNA Primers. The gaps are closed with the action of DNA Polymerase (adds complementary nucleotides to the gaps) and DNA Ligase (adds phosphate in the remaining gaps of the phosphate - sugar backbone). Each new double helix is consisted of one old and one new chain. This is what we call semiconservative replication. 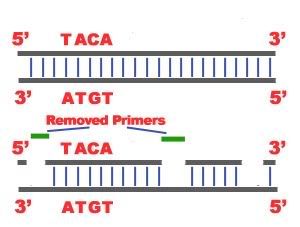 5) The last step of DNA Replication is the Termination. This process happens when the DNA Polymerase reaches to an end of the strands. We can easily understand that in the last section of the lagging strand, when the RNA primer is removed, it is not possible for the DNA Polymerase to seal the gap (because there is no primer). So, the end of the parental strand where the last primer binds isn't replicated. These ends of linear (chromosomal) DNA consists of noncoding DNA that contains repeat sequences and are called telomeres. As a result, a part of the telomere is removed in every cycle of DNA Replication.  6) The DNA Replication is not completed before a mechanism of repair fixes possible errors caused during the replication. Enzymes like nucleases remove the wrong nucleotides and the DNA Polymerase fills the gaps. |
| The Following User Says Thank You to AFRMS For This Useful Post: | ||
dr.atifrana (Tuesday, June 16, 2009) | ||
 |
«
Previous Thread
|
Next Thread
»
|
|
 Similar Threads
Similar Threads
|
||||
| Thread | Thread Starter | Forum | Replies | Last Post |
| Very Important : How to Prepare Study Notes | Shaa-Baaz | Tips and Experience Sharing | 5 | Sunday, May 21, 2017 08:30 PM |
| Effective Study Skills | Sureshlasi | Tips and Experience Sharing | 1 | Friday, November 16, 2007 09:28 AM |
| Regarding Notes | Anonymous84 | Tips and Experience Sharing | 1 | Wednesday, August 15, 2007 06:56 PM |












